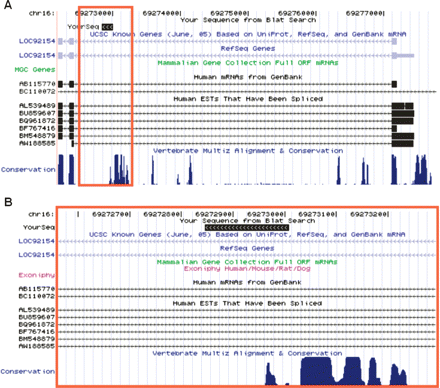
A novel AP on human chromosome 16 discovered by our computational prediction and experimental verification. B is an expanded view of the red rectangle region in A (images captured from the UCSC Genome Browser). A conserved promoter with a representative isoform NM_138383 (shown as LOC92154 above) was predicted to be an AP with an approximate log-likelihood ratio of +21.2, although aligned cDNAs and ESTs only supported a single promoter (BC110072 lacks any exonic overlap with this gene, and thus, is unlikely to represent an upstream AP). The top black box denoted by “YourSeq” represents the genomic alignment of a first exonic sequence found in our oligo-capped RACE reads. In B, the bottom rows indicate highly conserved blocks in the region immediately upstream of the detected first exon. The direction of transcription is from right to left.











