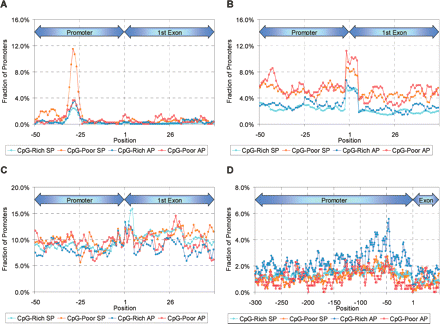
Positional distribution of TATA box motif, TATA (Butler and Kadonaga 2002) (A), initiator motif, YYANWYY (Butler and Kadonaga 2002) (B), downstream promoter element motif, RGWYV (Butler and Kadonaga 2002) (C), and CTCF binding site (combined count of CCCTCC [Filippova et al. 1996] and its complement) (D). For each nucleotide position, the number of promoters of a given type having a motif copy spanning that position is divided by the total number of promoters of that type. We used known promoters filtered as described in the Methods (Motif discovery and TRANSFAC search).











