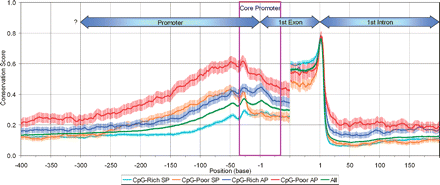
Average sequence conservation score (using UCSC 17-vertebrate alignment [Siepel et al. 2005]) at each nucleotide position in promoter, first exon, and first intron regions for each promoter type. Exon scores were computed for 50 exonic bases (or half the exon size, for exons <100 bp) from the 5′ or 3′ exon end. APs show on average higher conservation than SPs, and CpG-poor promoters higher conservation than CpG-rich promoters, over several hundred bases in the promoter and first intron, and in the 5′ half of first exon. Conservation patterns in the 3′ half of first exon largely reflect protein-coding constraints. The difference in sequence conservation becomes negligible further upstream of the TSS (which effectively eliminates the possibility that sequence-conservation differences near the TSS reflect large-scale variation in mutational rate rather than purifying selection). Uncertain TSS placement due to variable start sites, common in CpG-rich promoters, may cause some smearing of the conservation pattern for such promoters, but cannot by itself cause the overall weaker pattern. The boxed core promoter is bases −35 to +35 relative to the TSS.











