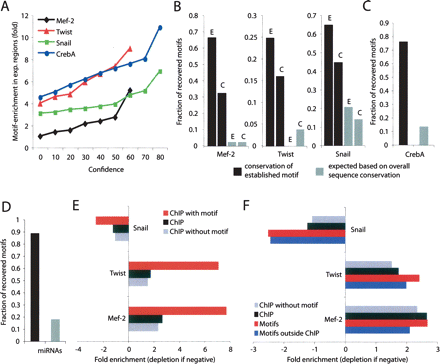
Conserved motif instances identify functional in vivo targets. Functional in vivo targets were determined for Mef-2, Twist, and Snail using ChIP-chip (Sandmann et al. 2006, 2007; Zeitlinger et al. 2007), and direct transcriptional targets were determined for CrebA using various assays (Abrams and Andrew 2005). (A) Increasing confidence values show increased enrichment for in vivo sites. Fold enrichment in functional in vivo sites (Y-axis) for conserved motif instances at varying confidence values (X-axis). Hypergeometric P-values for max fold enrichments are 4 × 10−11 for Mef-2, 2 × 10−6 for Twist, 3 × 10−10 for Snail, and 1 × 10−7 for CrebA. Increasing confidence levels selected functional in vivo sites with increased enrichment for all four regulators, showing that high conservation selects for functional motif instances (X = 0% shows the enrichment in the absence of comparative information, i.e., without requiring conservation). Curves are truncated when motifs do not reach the respective confidence levels. (B–D) High-sensitivity recovery of in vivo targets for TF and miRNA regulators. Fraction of motifs in bound regions recovered at 60% confidence (black bars), compared to the fraction expected given the overall conservation of the respective regions, as assessed by control motifs using the same BLS cutoff (gray; suggesting preferential conservation of the corresponding TF motif instances). (B) Recovery of ChIP-bound motifs, across all ChIP-bound regions (lableled “C”), and only those instances overlapping known enhancers (labeled “E”). Recovery rates show high sensitivity for TF motif instances, especially when these overlap enhancer regions. (C) Recovery of experimentally validated direct CrebA targets shows even higher sensitivity, likely due to the multiple lines of experimental evidence establishing them as direct targets. (D) miRNA recovery at 80% confidence is very high. (E) Nonconserved ChIP sites show reduced functional enrichments. Enrichment in promoter regions of muscle genes for motif instances of activators Twist and Mef-2, and depletion for motif instances of repressor Snail are reduced for ChIP-bound regions for which motif instances are not conserved, suggesting they may contain a higher fraction of nonfunctional sites. The enrichment/depletion is even weaker for ChIP-bound regions without motif instances (all enrichments are significant with P-values between 1.1 × 10−4 and 5.1 × 10−13 except those for Snail). (F) Conservation-inferred targets and ChIP-inferred targets show comparable functional enrichments. Conservation-inferred motif targets at 60% confidence (red; all P < 10−4) show higher muscle-gene enrichment/depletion than ChIP-inferred targets (black). Even outside ChIP-bound regions, conserved motifs show comparable enrichment and depletion (blue; all P < 5 × 10−3).











