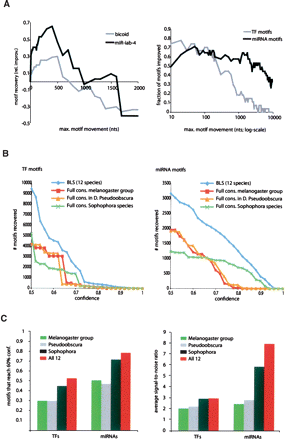
Discovery power for motif instance prediction. (A) Effect of tolerated motif movement. Number of recovered motif instances at 60% confidence for TF and miRNA motifs. (Left panel) For both TF motifs (gray: bicoid motif, VVVBTAATCC) and miRNA motifs (black: miR-iab-4 motif, GTATACG), instance recovery increases until an optimal window size (500 and 400 nucleotides, respectively) and then decreases for larger movements, suggesting that tolerating motif movements increases overall discovery power. (Right panel) Performance across all TF motifs (black) and all miRNA motifs (gray) shows improved recovery until windows of 300–500 nucleotides (for 60%–80% of motifs) but reduced performance for larger window sizes. Performance for individual examples (left panel) shows a sharper peak than the overall performance across all motifs (right panel), as different window sizes are optimal for different motifs. (B) BLS measure leads to increased sensitivity. Number of motif instances recovered (Y-axis) at each confidence value (X-axis) for transcription factor (TF) motifs (left panel) and miRNA motifs (right panel). The BLS measure applied to the 12 fly genomes (blue) recovers more motif instances at each confidence, as compared to approaches requiring motif presence in all compared species (“full” conservation), applied to the five melanogaster species (red), the pairwise comparison of D. melanogaster and D. pseudoobscura (yellow), or the nine Sophophora species (green). (C) Additional species lead to increased specificity. Two measures of discovery power for the BLS measure applied to the five melanogaster group species (green), a pairwise comparison of D. melanogaster and D. pseudoobscura (gray), the nine Sophophora species (black), and all 12 Drosophila species (red). (Left panel) More TF and miRNA motifs reach 60% confidence for increasing number of genomes at larger evolutionary distances. (Right panel) Increasing numbers of genomes at larger evolutionary distances also lead to increased signal-to-noise ratio, measured as the conservation level of real motifs vs. control motifs at the most stringent BLS cutoff.











