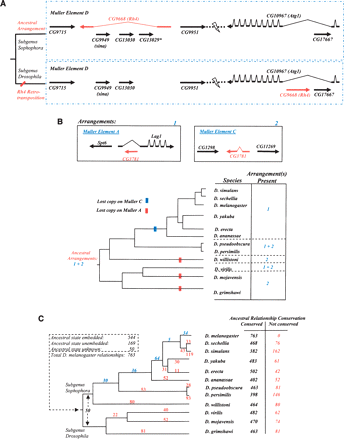
PRGs identified among embedded genes. (A) Embedded gene relationships. Such relationships are observed to be labile. This example illustrates how embedded relationships can change. The gene Rh4 (CG9668) has three embedded genes in D. melanogaster. The embedded relationship involving Rh4 and one gene representing sina (CG9949) or CG13030 (which are duplicated genes) was ancestral, based on conservation in the outgroup mosquitoes (Aedes aegypti, A. gambiae). Previous analysis had shown the movement of Rh4 (Neufeld et al. 1991) to a different location on the same chromosome arm (Muller Element D). Our analysis reveals the ancestral arrangement of these genes, the position of the Rh4 transposition event within the Drosophila phylogeny, and the creation of a new embedded relationship as a result of this relocation. Movement of Rh4 into an embedded position within an intron of Atg1 (CG10967) on the same arm is most likely the result of a retrotransposition event as the relocated Rh4 is intronless in its new location. The loss of the ancestral copy of Rh4 caused sina and CG13030 to be unembedded. The third embedded gene in Rh4, CG13029, is not found outside of the D. melanogaster–D. ananassae lineage. Gene extents are not to scale and intron-exon structure has been shown for Rh4 and Atg1 only. (B) Changes to an ancestral embedded relationship via differential loss of retrotransposed or parent gene. Because of a previous retrotransposition, duplicated genes paralogous to CG3781 are thought to be present in the last common ancestor to the genus Drosophila, with the relocated copy hypothesized to have arisen as an embedded copy on Muller A, originating from gene CG3781 on Muller C. Three independent losses of the gene copy on Muller Element A and one on Muller Element C appear to have occurred, although there exists the possibility that some of the Muller Element C losses might be due to the gene being absent in one or more of the assemblies. Gene order and orientation is depicted in the diagram: Intron and exon structure is not shown for all genes and gene extents are not to scale. (C) Differential conservation of embedded gene relationships. The phylogeny shows the distribution of the inferred positions of occurrence of the remaining nonancestral embedded relationships in the lineage leading to D. melanogaster (blue numbers). Loss of ancestral embedded relationships in non-melanogaster lineages are shown (red numbers) and totals for each species are presented. Such losses can occur as a result of gene movement or gene loss in a given lineage. Because gene prediction methods are more error prone in regions containing embedded genes, this analysis was based on the alternative Synpipe methodology (Bhutkar et al. 2006) not requiring gene models in the test species. Briefly, translations of the genomic regions of the other 11 species were assessed for the embedded or unembedded state using TBLASTN and the D. melanogaster protein set as queries (see Methods).











