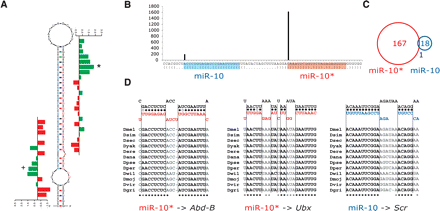
miR-10 and miR-10* target Hox genes. (A) SVM Z-scores indicate that miR-10 and miR-10* are both functional and that miR-10* is likely the major miRNA (green and red are positive and negative scores, respectively). (B) Cloning confirms that both sequences are expressed but that miR-10* is more abundant (306/9 in ovaries/testes and 1319/189 otherwise), consistent with the Z-scores. Shown is the number of total Solexa and 454 (Ruby et al. 2007) reads supporting each 5′ end position along the miRNA precursor hairpin (the more abundant miR-10* is shaded in red and miR-10 in blue). (C) Target genes of miR-10 and miR-10* are largely different. miR-10* targets 167 genes as compared to the 18 predicted for miR-10, with only one overlapping gene (Venn diagrams). (D) miR-10 and miR-10* both target Hox genes. miR-10* has highly conserved canonical sites in the Hox genes Abdominal-B (Abd-B) and Ultrabithorax (Ubx). miR-10 has a highly conserved compensatory target site in the Hox gene Sex combs reduced (Scr; Brennecke et al. 2005). In the absence of sequence-similar miRNAs, these sites argue that both sequences are functioning by regulating Hox genes. This relationship is similar to that of other Hox miRNAs in fly and mammals (Stark et al. 2003; Yekta et al. 2004).











