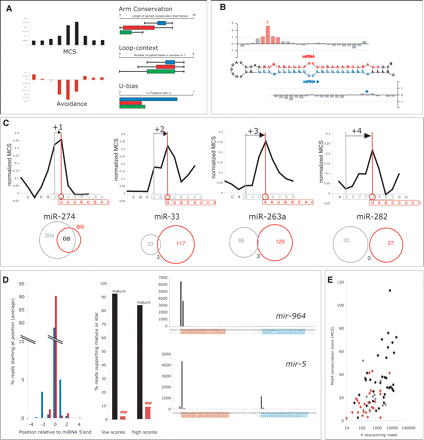
Properties of mature miRNAs. (A) Properties mature miRNA 5′ ends. 7-mers complementary to the start of mature miRNAs show a characteristic profile of 3′ UTR motif conservation scores (MCS) and avoidance in 3′ UTR of anti-target genes (normalized Z-scores averaged over nonredundant cloned miRNAs). miRNA 5′ ends are followed by a long stretch of perfect conservation that corresponds to the mature miRNA, and are in regions with constraint base-pairing; 78% of Drosophila miRNAs start with a uridine, whereas only ∼30% of all hairpin positions are uridine (boxes and whiskers denote 25 and 95 percentiles, for miRNA 5′ ends [blue], random arm positions [red], random hairpin positions [green]). (B) Combined properties accurately highlight the start of mature miRNAs. For an average miRNA hairpin, SVM Z-scores that combine above properties strongly highlight the start of the mature sequence (red, scores > 1), whereas they are low or negative (gray) throughout the remainder of the hairpin. Shown is the average Z-score over all nonredundant cloned Drosophila miRNAs along a representative hairpin (averages are calculated for each position after aligning all miRNAs and miRNA*s separately at their starts). (C) Correction of Drosophila miRNA annotation. Predicted and validated changes of mature miR-263a, miR-274, miR-282, and miR-33 (black lines denote the MCS profile for the new start sites; gray and red denote old and new sequences, respectively). These changes (between 1 and 4 nt) shifted the target spectrum of the miRNA drastically (Venn diagrams), such that the target genes of the annotated (gray circles) and corrected sequences (red circles) had generally little or no overlap. (D) miRNAs show alternate mature forms. miRNAs for which we correctly predict the 5′ end show more precise processing on average (90% of all reads supporting the mature 5′ end at x = 0; red). In contrast, the remaining miRNAs show a significant drop of the central peak with more reads coming from other positions (78%, P = 6 × 10−3; blue; left). The fraction of reads supporting the star sequence (red; shown is the median percentage across all cloned miRNAs) increases for high-scoring stars, while the reads supporting the mature miRNA (black) drops (middle). Number of reads from Ruby et al. (2007) supporting different 5′ end positions (mature and star sequences are shaded in red and blue, respectively; right). Novel miRNA mir-964 has two mature miRNAs at a ratio of 2:1: The major 5′ end is supported by 6457 (6393 from ovaries/testes) reads, but we predict position +1, which is supported by 3670 (3641 from ovaries/testes) reads. miR-5 and miR-5* are processed in a ratio of 4:1. (E) miRNA abundance correlates strongly with 3′ UTR motif conservation. Shown is the number of 454 sequencing reads (Ruby et al. 2007) for the dominant small RNA per hairpin and miRNA family (X-axis) versus the motif conservation score (MCS) of the corresponding 3′ UTR motif. A strong correlation between both measures is found with a Pearson correlation coefficient of 0.72. Colors denote Rfam miRNAs (previously cloned in black, others in gray) and novel validated miRNAs.











