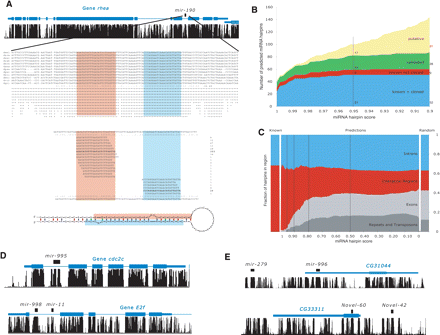
Novel Drosophila miRNAs. (A) Prediction and validation of miRNA mir-190. mir-190 (black) is predicted in the intron of the cytoskeleton anchor protein rhea (blue, UCSC browser screen-shot) in the direction of transcription, sequence alignment of mir-190 across 12 Drosophila genomes and conservation profile highlighting the mature miRNA (red) and the star sequence (blue). “.” and “()” denote unpaired and paired nucleotides according to Hofacker et al. (1994); experimental validation of mir-190 (total read counts from our and Ruby et al.’s [Ruby et al. 2007] sequencing shown to the right). Matching sequence reads show a characteristic pattern of processing with the total reads obtained for the miRNA and the star sequence, indicative of true Drosophila miRNAs. (B) Recovery of known novel miRNAs. Count of predicted miRNAs (Y-axis) at different score cutoffs (X-axis), for cloned (training set, blue), previously annotated but not cloned (red), novel and validated (green), and additional novel (yellow). At a conservative cutoff of 0.95, we recover 51 (85%) of cloned and nine previously annotated miRNAs among a total of 101 predictions. Of the 41 novel miRNAs, 24 (59%) are experimentally validated. A more lenient cutoff of 0.9 predicts 49 additional miRNAs, including one additional known miRNA and four novel miRNAs that are validated experimentally. (C) High-scoring hairpins are specific to introns and intergenic regions and exclude exons, repeats, and transposons. Shown are percentages for each region (Y-axis) for hairpin scores from 1.0 (best) to 0.0 (worst; X-axis). This is compared to the ratios obtained for random hairpins (Random) and known miRNAs (Known). For scores <0.8 the distribution of hairpins is indistinguishable from random, arguing that no further conserved hairpins can be expected at a reasonable frequency. (D) Examples of novel intronic 1 (four total) and clustered (six total) miRNAs. mir-995, that is in an intron of cdc2c and mir-998 is ∼500 nt from mir-11 in the intron of Ef2. (E) Novel miRNAs explain transcript of erroneously annotated genes. CG31044 and CG33311 are likely the precursor transcripts of mir-996, which is ∼2.5 kb from mir-279 (a member of the same family) and Novel-60, which is ∼1300 nt from Novel-42.











