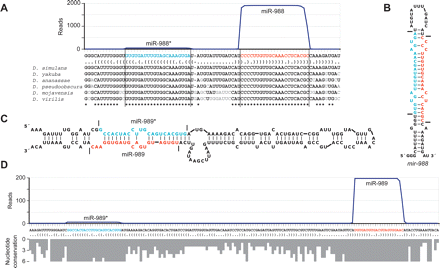
Newly identified miRNAs. (A) The sequence and bracket-notation secondary structure of the mir-988 hairpin, highlighting the miRNA (red) and the miRNA* (blue), with read abundance along the length of the sequence plotted above and orthologous hairpins aligned below; nonconserved nucleotides in gray (Drosophila Comparative Genome Sequencing and Analysis Consortium 2007a, b). Vertical lines indicate the inferred Drosha and Dicer cleavage sites. Analogous data for all newly identified D. melanogaster miRNAs are provided (Supplemental Table S2). (B) The predicted secondary structure of the mir-988 hairpin, colored as in A. Horizontal lines indicate the inferred Drosha and Dicer cleavage sites. (C) The unusually large hairpin of mir-989, colored as in A. (D) The sequence and bracket-notation secondary structure of the mir-989 hairpin, with coloring and read-abundance display as in A. Conservation across the length of the hairpin is shown below as a histogram, with bar depth indicating for each nucleotide the number of orthologs from the organisms shown in A with that nucleotide conserved.











