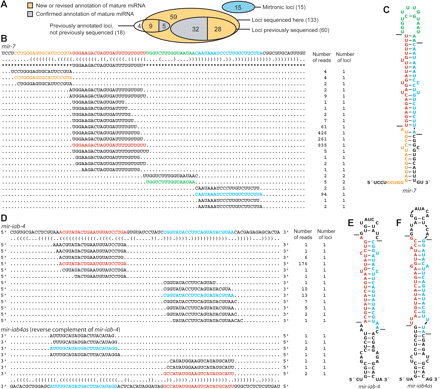
Correspondence between previously annotated miRNA hairpins and sequenced miRNAs. (A) Overlap between previously annotated miRNA hairpins and the total set of 133 hairpins of canonical miRNAs supported by our high-throughput sequencing (Supplemental Table S2). Mirtronic loci are described elsewhere (Ruby et al. 2007). (B) Small RNAs derived from the mir-7 hairpin. A portion of the mir-7 transcript is shown above its bracket-notation secondary structure, mature miRNA annotation from miRBase v8.1 (Griffiths-Jones 2004) flanked by asterisks, and sequences from the present study. For each sequence, the number of reads giving rise to that sequence and the number of loci to which the sequence maps in the D. melanogaster genome are shown on the right. Highlighted are the most abundant sequences corresponding to the miRNA (red), miRNA* (blue), intervening loop (green), and fragment flanking the 5′ Drosha cleavage site (orange) (Supplemental Text). Analogous data for all previously annotated D. melanogaster miRNAs are provided (Supplemental Table S2). (C) The predicted hairpin structure of the mir-7 hairpin, colored as in B. Lines indicate inferred Drosha and Dicer cleavage sites. (D) Small RNAs derived from the mir-iab-4 and mir-iab4as hairpins, displayed as in B. (E) The predicted secondary structure of the sense mir-iab-4 hairpin precursor, formatted as in C. (F) The predicted secondary structure of the mir-iab-4 reverse complement, mir-iab4as, formatted as in C.











