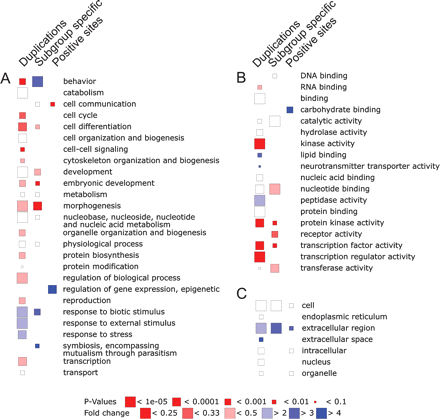
Different classes of rapidly evolving genes. Duplicated genes are often involved in adaptive functions such as responses to external stimuli, whereas they are under-represented in transcription factors and regulatory genes. Shown are over-/under-represented GOSlim categories of D. melanogaster genes present in clusters containing gene duplications (n = 1126) (A), without detectable orthologs in species further diverged than D. yakuba and D. erecta (n = 795) (B), and with sites predicted to have been subject to positive selection (n = 121) (C). The size of the box represents the P value of the over/under-representation while the fold over-/under-presentation is indicated by the color of the box (see scale at bottom). False-positive predictions arising from the application of multiple tests were controlled using a false discovery rate of 0.05.











