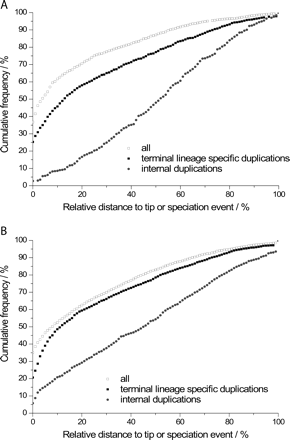
Approximately 20%–30% of all gene duplications have been very recent, whereas duplications inferred to have been more ancient occurred less frequently and more uniformly. Duplication events have been dated by the synonymous substitution rate dS and normalized by the overall height of each gene tree (open squares) or by the corresponding branch length (solid squares) of the species tree after reconciliation with the gene tree. The latter have been aggregated over all internal or terminal lineage branches, respectively. A similar picture emerges when considering each branch separately (Supplemental Figs. S9 and S10). Duplications from D. melanogaster subgroup only rooted with D. pseudoobscura and/or D. persimilis sequences (A) or all 12 species (B). Among 13,132 clusters, 5851 had the full species complement and there were 1853 clusters with 1305 internal and 3794 lineage-specific duplications.











