Genes lined to neurodegenerative and developmental diseases are enriched for variable intragenic tandem repeats
Click on table to view larger version.
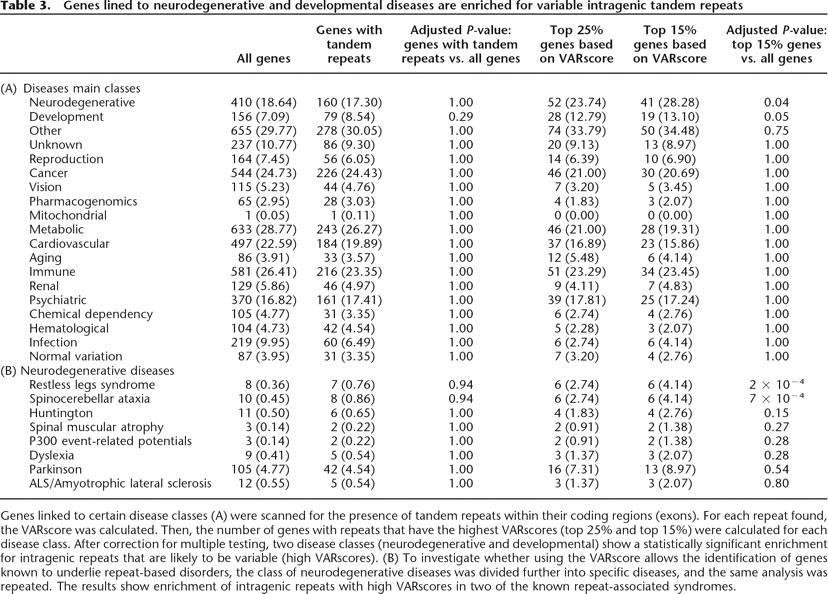
Genes linked to certain disease classes (A) were scanned for the presence of tandem repeats within their coding regions (exons). For each repeat found, the VARscore was calculated. Then, the number of genes with repeats that have the highest VARscores (top 25% and top 15%) were calculated for each disease class. After correction for multiple testing, two disease classes (neurodegenerative and developmental) show a statistically significant enrichment for intragenic repeats that are likely to be variable (high VARscores). (B) To investigate whether using the VARscore allows the identification of genes known to underlie repeat-based disorders, the class of neurodegenerative diseases was divided further into specific diseases, and the same analysis was repeated. The results show enrichment of intragenic repeats with high VARscores in two of the known repeat-associated syndromes.











