Specific classes of human genes show enrichment for variable intragenic repeats
Click on table to view larger version.
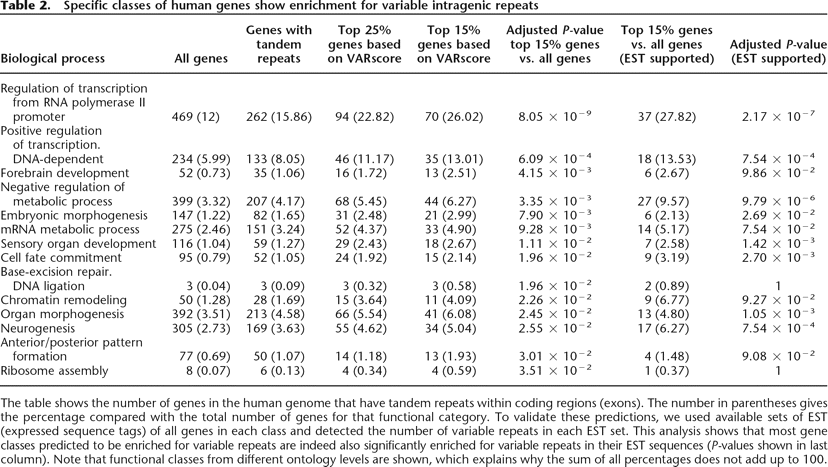
The table shows the number of genes in the human genome that have tandem repeats within coding regions (exons). The number in parentheses gives the percentage compared with the total number of genes for that functional category. To validate these predictions, we used available sets of EST (expressed sequence tags) of all genes in each class and detected the number of variable repeats in each EST set. This analysis shows that most gene classes predicted to be enriched for variable repeats are indeed also significantly enriched for variable repeats in their EST sequences (P-values shown in last column). Note that functional classes from different ontology levels are shown, which explains why the sum of all percentages does not add up to 100.











