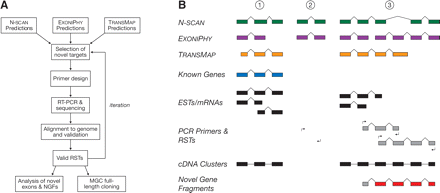
(A) Flowchart for computational exon discovery (CED). Beginning with three sets of gene predictions, candidate novel genes are tested for evidence of expression and splicing in several rounds of candidate selection, RT–PCR amplification, and sequencing. The result is a large set of EST-like sequences, called RSTs, that provided supporting evidence for novel protein-coding exons, but do not define full-length transcripts. (B) Illustration of CED. Gene 1 is known and well-supported by public cDNA sequences, so overlapping gene predictions are ignored. Predicted gene 2 appears to be novel and is selected for RT–PCR validation, but the validation experiment fails. Predicted gene 3 also appears to be novel and is tested by two RT–PCR experiments, both of which produce valid RSTs (“hits”). The first experiment validates the TRANSMAP prediction, and the second validates the N-SCAN prediction and one of two Exoniphy predictions. A cDNA cluster is constructed to summarize each set of overlapping cDNAs (including RSTs), and a novel gene fragment (NGF) is constructed by merging the two RSTs that support novel exons (NEs; in red).











