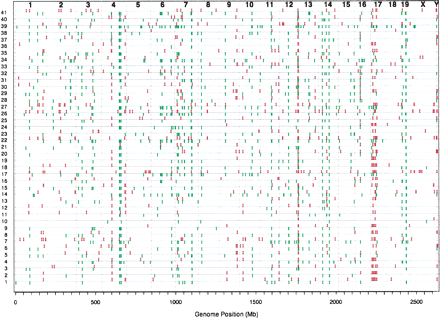
All mouse CNVs. Predicted CNVs for each inbred mouse strain are displayed based on their genomic position. Amplifications are shown in red above the baseline for each strain, deletions are shown in green below. The strains are: (1) 129X1/SvJ; (2) 129S1/SvImJ; (3) AKR/J; (4) A/J; (5) BTBR T+ tf/J; (6) BUB/BnJ; (7) BALB/cJ; (8) C3H/HeJ; (9) C57BLKS/J; (10) C57BL/10J; (11) C57BR/cdJ; (12) C57L/J; (13) C58/J; (14) CAST/EiJ; (15) CBA/J; (16) CE/J; (17) CZECHII/EiJ; (18) DBA/1J; (19) DBA/2J; (20) FVB/Ntac; (21) I/LnJ; (22) JF1/Ms; (23) KK/HIJ; (24) LP/J; (25) MA/MyJ; (26) MOLF/EiJ; (27) MSM/Ms; (28) NOD/LtJ; (29) NON/LtJ; (30) NZB/BINJ; (31) NZW/LacJ; (32) PERA/EiJ; (33) PL/J; (34) PWK/PhJ; (35) RIIIS/J; (36) SEA/GnJ; (37) SJL/J; (38) SM/J; (39) SPRET/EiJ; (40) SWR/J; (41) WSB/EiJ.











