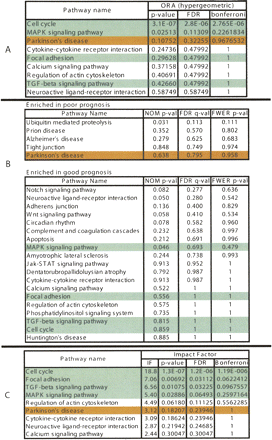
A comparison between the results of the classical (ORA) probabilistic approach (A), GSEA (B), and the results of the pathway impact analysis (C) for a set of genes associated with poor prognosis in breast cancer. The pathways marked with green are well supported by the existing literature. The ones in red are unlikely to be related. After correcting for multiple comparisons, GSEA fails to identify any pathway as significantly impacted in this condition at any of the usual significance levels (1%, 5%, or 10%). The hypergeometric model pinpoints cell cycle as the only significant pathway. Relevant pathways such as focal adhesion, TGF-beta signaling, and MAPK do not appear as significant from a hypergeometric point of view. While agreeing on the cell cycle, the impact analysis also identifies the three other relevant pathways as significant at the 5% level.











