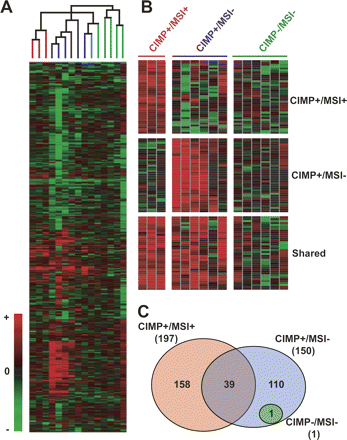
Global methylation analysis of colorectal carcinomas by MCAM. (A) Cluster analysis of MCAM data in primary colorectal carcinomas. A total of 4588 probe data was used for unsupervised hierarchical clustering analysis of 15 sample pairs (tumor compared to normal appearing mucosa), resulting in a classification that matched the known CIMP/MSI status of these samples. The terminal branches are color-coded to represent the CIMP/MSI status of the tumor sample: (red) CIMP+/MSI+; (blue) CIMP+/MSI−; (green) CIMP−/MSI−. (B) Probe clusters, which classifies CIMP+/MSI+ and CIMP+/MSI− samples apart from each other, and CIMP+ in general (shared group) from CIMP− samples. (C) Venn diagram of concordant probe methylation (at least 60% of samples methylated) for each CIMP/MSI group. The total number of methylated probes in each group is shown in parenthesis.











