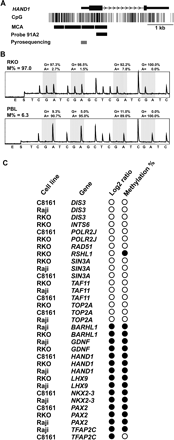
Validation of methylation status of selected genes in cancer cell lines. (A) Representation of one identified methylated gene by MCAM. One of several possible MCA fragments (delimited by CCCGGG sequences) close to the transcription start site of the HAND1 gene overlaps with the DNA probe 91A2, allowing the investigation of this fragment in the microarray platform. Note that the remaining MCA fragments do not overlap additional probes, being therefore not investigated in this system but potentially investigated in other arrays with different probe collections. (Gray bar) The promoter region investigated by bisulfite PCR followed by pyrosequencing. (B) Representative pyrograms for HAND1. Four CpG sites close to the transcription start site were pyrosequenced, and a consistent pattern of high levels of methylation was observed for all of them in RKO, while in peripheral blood lymphocyte the methylation values were low, consistent with the MCAM results. The methylation density is presented in the top of each pyrogram as the averaged methylation of the four studied CpG sites. (C) Graphic representation of log2 ratio and methylation analysis by bisulfite-PCR results for selected genes for validation of MCAM. (Black circles) Hypermethylation in tumor (as determined as log2 ratio ≥ 1.3 in MCAM and methylation density >10% by bisulfite-PCR followed by pyrosequencing); (white circles) lack of methylation. Note the high concordance of methylation results between techniques.











