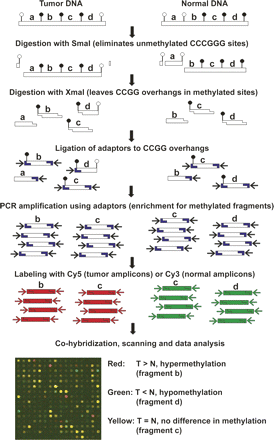
Schematic diagram of the MCAM method. Enrichment for methylated DNA and reduction of genome complexity is achieved by serial digestion with SmaI (methylation sensitive) and XmaI (methylation insensitive) restriction enzymes, followed by ligation of adaptors and PCR amplification. The resulting amplicons, representative of the methylated fraction of tumor and normal cells, are labeled and cohybridized in a microarray platform. Image acquisition and data analysis allow identification of methylated and nonmethylated genes by comparing intensity values of Cy5 and Cy3 dyes for each pair of tumor and control samples.











