Results for chimpanzee species
Click on table to view larger version.
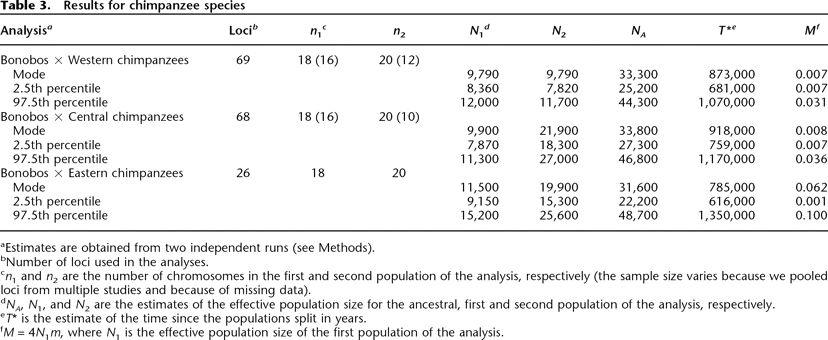
aEstimates are obtained from two independent runs (see Methods).
bNumber of loci used in the analyses.
cn1 and n2 are the number of chromosomes in the first and second population of the analysis, respectively (the sample size varies because we pooled loci from multiple studies and because of missing data).
dNA, N1, and N2 are the estimates of the effective population size for the ancestral, first and second population of the analysis, respectively.
eT* is the estimate of the time since the populations split in years.
fM = 4N1m, where N1 is the effective population size of the first population of the analysis.











