Performance of MIMAR when detecting gene flow
Click on table to view larger version.
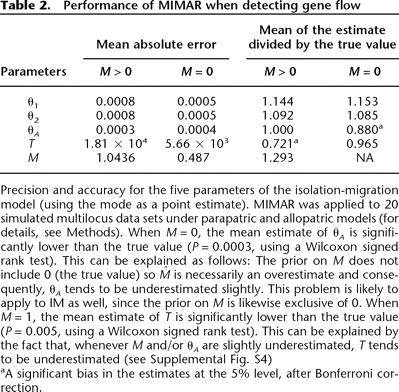
Precision and accuracy for the five parameters of the isolation-migration model (using the mode as a point estimate). MIMAR
was applied to 20 simulated multilocus data sets under parapatric and allopatric models (for details, see Methods). When M = 0, the mean estimate of θA is significantly lower than the true value (P = 0.0003, using a Wilcoxon signed rank test). This can be explained as follows: The prior on M does not include 0 (the true value) so 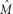 is necessarily an overestimate and consequently, θA tends to be underestimated slightly. This problem is likely to apply to IM as well, since the prior on M is likewise exclusive of 0. When M = 1, the mean estimate of T is significantly lower than the true value (P = 0.005, using a Wilcoxon signed rank test). This can be explained by the fact that, whenever M and/or θA are slightly underestimated, T tends to be underestimated (see Supplemental Fig. S4)
is necessarily an overestimate and consequently, θA tends to be underestimated slightly. This problem is likely to apply to IM as well, since the prior on M is likewise exclusive of 0. When M = 1, the mean estimate of T is significantly lower than the true value (P = 0.005, using a Wilcoxon signed rank test). This can be explained by the fact that, whenever M and/or θA are slightly underestimated, T tends to be underestimated (see Supplemental Fig. S4)
aA significant bias in the estimates at the 5% level, after Bonferroni correction.











