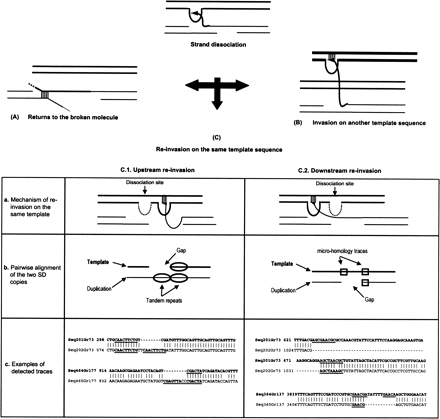
Re-invasion processes and signatures generated after strand dissociation. Template sequence is represented in bold. After the dissociation of the strand, the newly synthesized strand can return to the broken molecule (A), re-invade another genomic region (B), or re-invade the same template (C). In this latter case, traces are expected to be observed on the pairwise alignment of the template with the duplication sequence. For strand re-invasion, a microhomology is required (see text). (C.1) In the upstream re-invasion case, the same genomic region is used as template twice (a), generating a tandem repeat within the newly synthesized strand, although the template remains intact. The pairwise alignment would thus exhibit a gap on the template sequence associated with two tandem repeats of the duplication with only one repeat conserved (b, c). Microhomology traces could not be distinguished from the two tandem repeats because they overlap with the tandem repeats. (C.2) In the downstream re-invasion, the template jump would be visible through a gap on the duplication corresponding to the template region not synthesized (a). This gap would be associated with the traces of the re-invasion. They correspond to two small repeats—i.e., microhomology traces: one on one side of the gap and the other within the gap on the opposite side (b,c). In examples of traces, illustrated in c, tandem repeats and microhomology traces are underlined.











