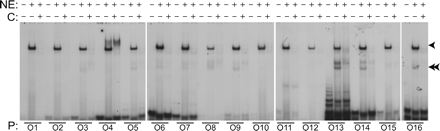
Nuclear protein binding to predicted novel (N1) motifs. Electromobility gel shift assays were performed to assess the ability of the motifs to bind proteins. Oligonucleotides (O1–O16) representing motifs (mouse) identified within the cartilage gene set were radiolabeled with [32P]ATP, incubated with nuclear extracts (NE) from rat chondrosarcoma cells, and the protein-binding moieties were separated by SDS-PAGE (4%). In some reactions, additional unlabeled oligonucleotide (C) was also added at 100× to evaluate specificity binding to the motifs. The presence or absence of nuclear extract (NE) and unlabeled oligonucleotides (C) is indicated in the rows corresponding to the labels by + and −, respectively. The radiolabeled oligonucleotides used in each reaction are indicated by lines above the numbered oligonucleotide from Table 2. Predominant protein-binding complexes are indicated by single and double arrowheads.











