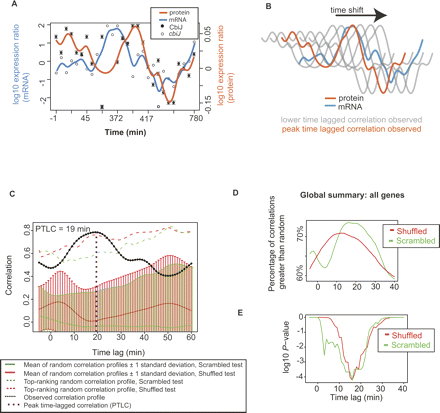
Time lags between transcription and translation were calculated for individual genes and for the global data set. (A) Interpolated profiles for an example gene. Quantitative iTRAQ protein data (log10 ratios; ●) and mRNA levels (log10 ratios; ○) are shown for CbiJ, the precorrin 3-methylase of the cobalamin biosynthesis pathway. Error bars represent the standard error in all peptide quantitations. Cubic spline interpolated profiles for the protein data (orange line), and mRNA (blue line) are overlaid (see legend). The X-axis represents time post-shift to high oxygen in the first replicate chemostat experiment (see also Fig. 1; Table 1). (B) Schematic of time shifting strategy for an example operon. Protein profiles such as those shown in A (orange) are shifted relative to interpolated mRNA profiles (blue) and the time lag Δt corresponding to the maximum correlation is recorded (peak time lagged correlation; PTLC). Gray lines represent protein profiles at less-than-optimum time lags, and the position at which the orange line (protein) overlaps the blue line (mRNA) represents the PTLC position. (C) Time-lagged correlation profile for cbiJ is significantly better than random. Permutation tests were performed whereby 100 different scrambled (green, with standard errors 1-σ from mean) and shuffled (red, with standard errors) mRNA profiles (see Methods) were subjected to the same time-lagged correlation comparison with the CbiJ protein data (as depicted in A and B) and compared with the true TLC for cbiJ (black dotted line). The most significant difference between the cbiJ profile and random was at Δt = 19 min (purple dotted line; P < 0.01). (D) The mean, global time lag for all genes occurs at ∼16 min. Approximately 75% of time lag correlations for all genes at this time lag are better than random, whereas only ∼60% of time lags are greater than random when no time lag is considered (Δt = 0). (E) The difference between these global percentages is significant for both types of permutation test (P ∼ 10−4), especially near Δt = 16 min. A and C are shown for several other genes and operons in Supplemental Figures 7 and 8, respectively.











