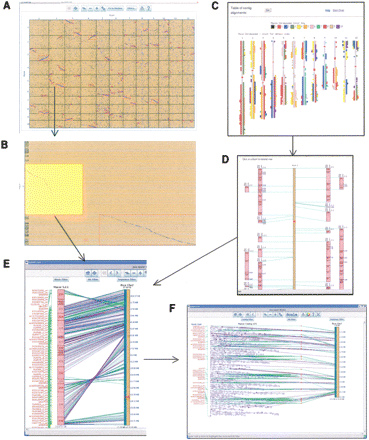
The SyMAP displays. (A) Java dot plot of two whole genomes, where the vertical axis is the FPC contigs and the horizontal axis is the sequenced genome. Each square represents the anchors shared by the two chromosomes. Clicking on a square zooms in on it. (B) From the square, a region can be selected which turns it yellow. Clicking a yellow region displays the close-up shown in E. (C) CGI/Perl display of the blocks aligned to the sequenced chromosomes. The blocks are color coded to indicate what FPC chromosome they are from. Clicking on a sequenced chromosome will show the close-up. (D) CGI/Perl display of the close-up of a sequenced chromosome, where the numbers in the small rectangles are contig numbers. Selecting a block will display the close-up. (E) Java close-up of the block. Purple lines are BESs. Green lines are markers. Select a contig to show the close-up. (F) Java close-up of the contig. The displays E and F have filters for the three tracks. The Block and Contig filters allow marker and clone names to be filtered on different properties. The Hit filter allows the lines between the anchor points to be filtered on different properties. The sequence filter allows the annotation, framework markers, and gaps to be made visible/invisible.











