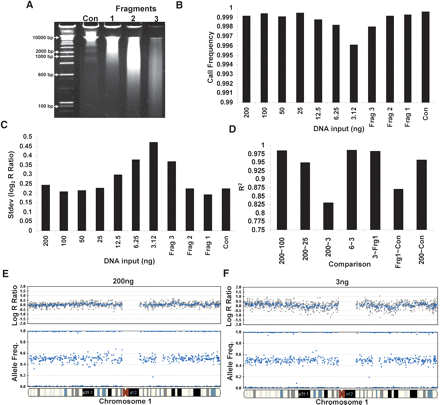
Effect of gDNA quantity and fragmentation on SNP-CGH data. SNP-CGH data quality as a function of the quantity and fragmentation length of gDNA in the amplification reactions was tested using a multisample 10K BeadChip format. (A) Various lengths of fragmented DNA were used as starting input for the whole-genome amplification reaction (Fragments 1, 2, and 3). (B) The call rate is relatively insensitive to input amount across the entire range of 200 ng to 3 ng or fragment length. The call rates for input DNA ranging from 25 to 200 ng were all above 0.999, and the call rates for the 3–12.5 ng were all above 0.996. (C) Genomic DNA was titrated from the standard 1× input (200 ng in a one-quarter scale reaction) down to 1/64th input (3 ng in a one-quarter scale reaction) as well as Fragments 1, 2, and 3. As the levels of input DNA decreased, the variability in the log R ratio noticeably increased, whereas the allelic ratio was relatively insensitive to input amounts. (D) The R 2 correlation between samples remains high when similar amounts of gDNA are used for input regardless if it falls into either high or low levels; however, the R 2 decreases dramatically between inputs that differ substantially in amount. (E) An example genome profile from chromosome 1 showing both the log R ratio and AF from the sample using 200 ng of input DNA. (F) The same plot as in E but with 3 ng of input DNA. Notice the slight increase in log R ratio variability with the lower amount of DNA input.











