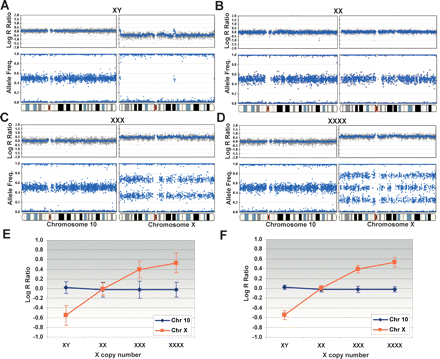
X-copy cell lines as a model system. Single copy deletions, monoallelic duplications (trisomies), and amplifications are detected on the Human-1 (109K) BeadChip using cell lines with one to four X chromosomes. All plots are shown juxtaposed to normal genome profiles from chromosome 10. (A) In XY, the presence of a single X chromosome is shown as a decrease in the log2 R ratio from ∼0to −0.55, and in the AF plot where the heterozygous state completely collapses to the homozygous axis. Note the pseudoautosomal regions on the Y chromosome. (B) In XX, the presence of the expected two copies of the X chromosome show no deflection in the log R ratio (∼0), and the heterozygotes are clustered around +0.5. (C) In XXX, the log2 R ratio increases from ∼0 to +0.395, and the heterozygous state splits into two clusters, one located at 0.67 (2:1 ratio) and the other at 0.33 (1:2 ratio). (D) In XXXX, the log2 R ratio increases to +0.53, and the heterozygous state is divided into three populations (0.25, 0.51, and 0.76) with allelic ratio of 3:1, 2:2, and 1:3, respectively. (E) The response of the log R ratio for both the X chromosome and chromosome 10 for each X-copy number cell line. The log R ratio increases with increasing X-copy number for the X chromosome but not for chromosome 10. The corresponding standard deviation is shown for each data point. (F) The log R ratio response calculated for the X-copy cell lines with a 10-SNP moving average. Note that the standard deviation has been significantly reduced. For all genomic profiles, the blue line indicates a 500-kb moving median for the Human-1 (109K) BeadChip. The complete X-copy cell line data set, cluster information, and receiver operator plots are available in the Supplemental material.











