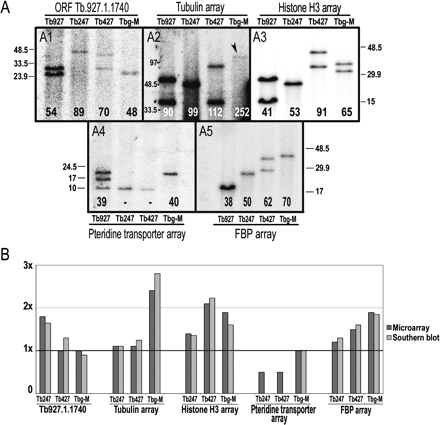
Validation of data derived from hybridization to the Tb927-chrIa tiling path genomic microarray. (A) Hybridization of five gene-specific probes to Southern-blotted genomic fragments containing intact tandem arrays. Numbers at the bottom of each lane indicate (in kb) total DNA content of each array in each genome, estimated from these gels. Two bands in the same lane indicate that the array varies in length in the chrI homologs. The 10-kb band in panel 4 probably is due to cross-hybridization with a similar sequence in a different chromosome, and therefore has not been included in the final length for the chrI region containing pteridine transporter genes. DNA size markers are in kb. (B) Comparison between microarray results and Southern blot results. y-axis represents the amount of DNA compared with Tb927. Value of 1× in the y-axis indicates equal DNA content compared with Tb927.











