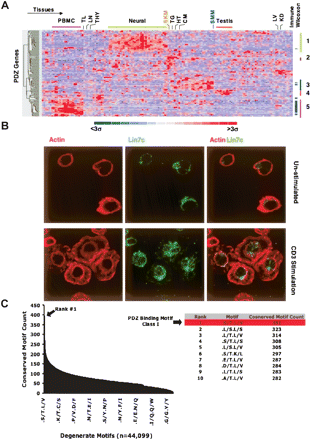
Systematic analysis of PDZ expression profiles and carboxy-terminal motifs. (A) mRNA expression profiles visualized across 79 tissue/cell types performed in duplicate from the human GNF compendium for 107 PDZ genes (Su et al. 2004). Genes and tissues were hierarchically clustered and visualized using dCHIP (Schadt et al. 2001). Selected samples aggregated into common tissue types are described at the top of the panel. (PBMC) Peripheral blood mononuclear cells, (TN) tonsil, (LN) lymph node, (THY) thymus, (SKM) skeletal muscle, (TG) tongue, (HT) heart, (CM) cardiac myocyte, (SMM) smooth muscle, (LV) liver, (KD) kidney. Four non-immune co-expression clusters among the various tissues are designated with corresponding genes in each cluster found in Supplemental Table 1. PDZ genes significantly enriched in the immune system (P < 0.05) as found by the Wilcoxon rank sum test are denoted with a tick mark on the right. (B) Endogenous expression of LIN7C in lymphocytes was confirmed by immunoflourescence in unstimulated or CD3-stimulated human Jurkat T cells. (C) The number of conserved instances for each of the observed 44,099 twofold generate motifs at positions −2 and 0 in the carboxyl terminus among the 11,044 aligned genes between human, dog, rat, and mouse. (Table) The top 10 ranked degenerate motifs showing that the Class I PDZ motif (–X-[S/T]-X-[L/V]-COOH) is the most conserved motif.











