Summary of unstable F-box-FTH and MATH-BTB trees
Click on table to view larger version.
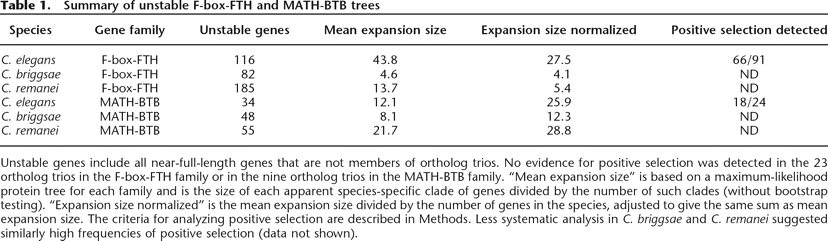
Unstable genes include all near-full-length genes that are not members of ortholog trios. No evidence for positive selection was detected in the 23 ortholog trios in the F-box-FTH family or in the nine ortholog trios in the MATH-BTB family. “Mean expansion size” is based on a maximum-likelihood protein tree for each family and is the size of each apparent species-specific clade of genes divided by the number of such clades (without bootstrap testing). “Expansion size normalized” is the mean expansion size divided by the number of genes in the species, adjusted to give the same sum as mean expansion size. The criteria for analyzing positive selection are described in Methods. Less systematic analysis in C. briggsae and C. remanei suggested similarly high frequencies of positive selection (data not shown).











