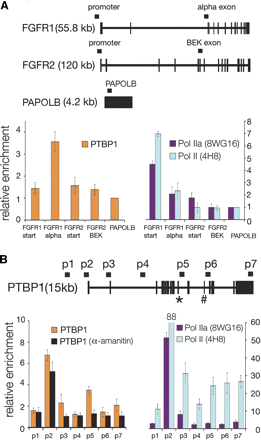
PTBP1 and RNA polymerase II are enriched at sites within transcribed genes. (A) Schematic representation of the FGFR1, FGFR2, and PAPOLB genes with primers depicted above genes; thick segments represent exons, and thin segments represent introns. PCR analysis of ChIP’s targeting PTBP1, promoter associated RNA polymerase II (Pol IIa, 8WG16), and total RNA polymerase II (non-phosphospecific Pol II, 4H8). Error bars represent the standard deviation calculated from three to seven biological repeats. (B) PTBP1, Pol IIa, and Poll II are enriched at PTBP1’s promoter and alternative exons. Starting upstream of the transcription start site, eight primer sets (denoted p1–p7) were designed to target sites across PTBP1. Alternative exons include exon 9 (*) and exon 11 (#). HeLa cells were grown in 40 μg/mL a-amanitin, and then cross-linked and sonicated for the α-amanitin ChIP experiments.











