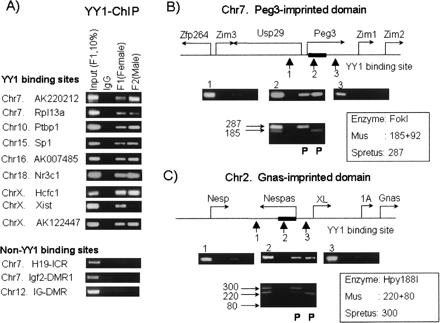
In vivo YY1 binding to other clustered regions. YY1 binding was tested on other clustered YY1 binding sites listed in Table 1. This series of ChIP analyses were also performed on three non-YY1 binding sites as negative control loci, including H19-ICR, Igf2-DMR1, and IG-DMR. The amplified PCR products of each cluster are shown in the following format: 10% of the Input DNA from F1 (lane 1), with pre-immune serum (lane 2), F1 brain (lane 3), and F2 brain (lane 4) with anti-YY1 antibody (A). We determined the allelic origin of the YY1-immunoprecipitated DNAs from the clustered site in Peg3-DMR (B) and Nespas-DMR (C). This analysis used sequence polymorphisms detected between the two parental species of our hybrid mice, F1 and F2. The restriction enzymes differentiating these sequence polymorphisms are shown with the estimated sizes of digested PCR products for each species. Two separate restriction enzyme digestions clearly demonstrated the presence of two alleles in the Input DNAs of F1 (lane 1). However, the YY1-immunoprecipitated DNAs at both Peg3-DMR and Nespas-DMR were mainly derived from the paternal allele (lanes 3 and 4).











