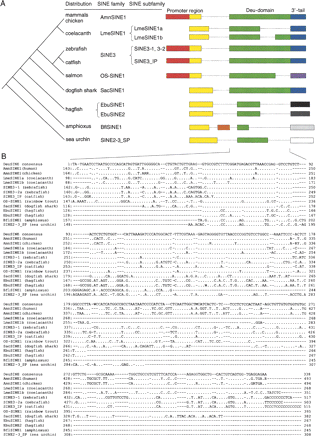
The structure and distribution of DeuSINEs. (A) Schematic representation of DeuSINEs and the known phylogenetic relationships between the host species. All of these SINEs were newly characterized in this study except for zebrafish SINE3 and sea urchin SINE2-3_SP. Green boxes represent common sequences among DeuSINEs, and an alignment of the region is shown in B. Yellow and red boxes denote promoter regions derived from tRNA (see Fig. 2A) and 5S rRNA (Fig. 2B), respectively. The regions denoted by yellow boxes in 5S rRNA-derived SINEs (AmnSINE1, SINE3, SINE3_IP and OS-SINE1) are similar to tRNA-derived promoter regions of LmeSINE1 (see Fig. 3 for details). Blue boxes represent 3′-tails of SINEs that are similar to that of zebrafish CR1-4_DR LINE (Fig. 4A), whereas the purple box is similar to that of the rainbow trout RSg-1 LINE (Fig. 4B). The 3′-tail sequences of EbuSINE1, EbuSINE2, BflSINE1, and SINE2-3_SP are distinct and of unknown origin. (B) Alignment of the common central DeuSINE sequences (Deu-domain; green boxes in A) in different host organisms. The SINE3-1 and the SINE3-2a sequences represent two subfamilies of the zebrafish SINE3 family. The top sequence is a consensus of the DeuSINE domain. Dots indicate nucleotides identical to those in the consensus sequence, and dashes indicate gaps inserted to improve the alignment.











