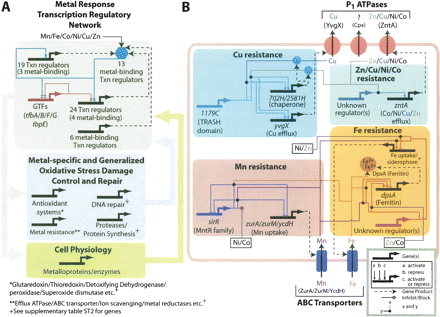
A systems-level model for transition metal stress response of Halobacterium NRC-1 (see text for details). (A) Transcription regulatory control of metal detoxification, oxidative stress, and general physiology in response to transitions metals. While most of the regulatory influences indicated were deciphered on the basis of mRNA level changes and putative functions, the regulatory control imposed by the GTFs were experimentally measured with ChIP-chip (M.T. Facciotti, M. Pan, A. Kaur, M. Vuthoori, D.J. Reiss, R. Bonneau, P. Shannon, S. Donahoe, L. Hood, and N.S. Baliga, in prep.). The blue and green watermark arrows indicate feedback signals. (B) Key functional and regulatory aspects of metal-resistance mechanisms. Regulatory control mediated by VNG1179C and SirR was verified by both microarray analysis as well as phenotypic assays on in-frame gene-deletion strains. Likewise, efflux of metal ions by YvgX and ZntA was also verified through phenotypic assays on gene deletion strains. Up-regulation of siderophore biosynthesis genes by Mn(II) was verified to be due to simulation of Fe-deficiency (see text). Finally, the role of the ferritin DpsA in Fe resistance and some aspects of regulation have been previously characterized (Reindel et al. 2002, 2005; Zeth et al. 2004). See inset key for interpreting the circuit.











