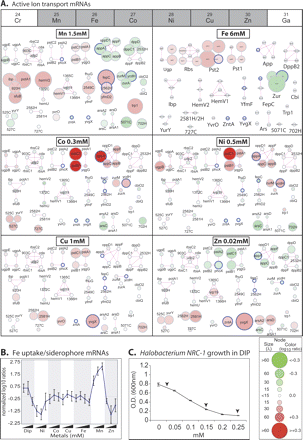
Responses of active transport and siderophore biosynthesis genes to the six metals and 2,2′ dipyridyl (DIP). (A) mRNA level changes in key transporters and genes in their putative operons. The genes are individually labeled at top, left and at top, right; transport systems are labeled as follows: Ugp: glycerol phosphate transport; Rbs: ribose transport; Pst1 and Pst2: PO42- transport; App and DppB2: peptide transport; Ibp, HemV2, HemV1, YfmC and FepC: Fe transport; Zur: Mn uptake; Cbi: Co uptake; YurY and YvrO: ABC transporters of unknown specificity; 2581H and 702H: putative Cu chaperones, (Ars) As resistance; 5071C: sugar transporter. Circular nodes represent genes of putative transport systems (Supplemental Table 2). Node coloring (red for up and green for down) indicates mRNA level change at 300 min in 1.5 mM Mn(II), 6 mM Fe(II), 0.3 mM Co(II), 0.5 mM Ni(II), 1 mM Cu(II), and 0.02 mM Zn(II)—organized in order of their position in the periodic table (top). Node size is proportional to λ value, a measure of the statistical significance of change (see inset key). Edges (lines) connecting the nodes indicate operon-like organization. (B) mRNA level changes in 12 putative siderophore biosynthesis and Fe-uptake genes in DIP and all six metals. (C) Halobacterium NRC-1 growth (OD600) at 27 h in increasing concentrations of the Fe-specific chelator DIP. Arrowheads indicate concentrations used for microarray analysis.











