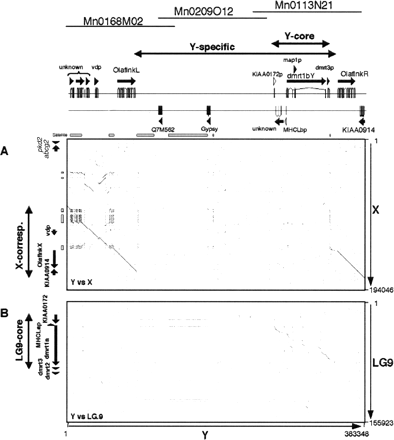
Comparison of the Y-chromosomal sequence to the X-chromosomal and LG9 sequences. Genes were identified in the sequenced regions of the sex chromosomes and LG9, represented by arrows along the dot plots. Sequences were compared using the Dotter software. Numbers on the right or at the bottom show the length of the sequences in base pairs. The two-headed arrows indicate the subregions (see legend for Fig. 2). The Y-chromosomal sequence region is compared against the X-chromosomal sequence region in A, and to LG9 in B. (A) The X and Y sequences are aligned at the left and right regions, but not in the middle of the Y region. The Olaflnk genes are delimiting the regions with homology. The sequence of OlaflnkR and the region to the right is aligned without gaps to the X sequence, whereas there are gaps in the alignment left to the OlaflnkL gene. There are also clusters of repetitive sequences in this region. Note that there are two stretches of sequence in this region that are inverted in the X sequence (depicted as two short lines running from left to right, upward). (B) The LG9 sequence cannot be aligned to most of the Y region, but there are some stretches of similarity between the sequences in the dmrt1bY and dmrt3p region.











