Differential tissue expression for X. laevis paralogs from EST information (top 20 genes)
Click on table to view larger version.
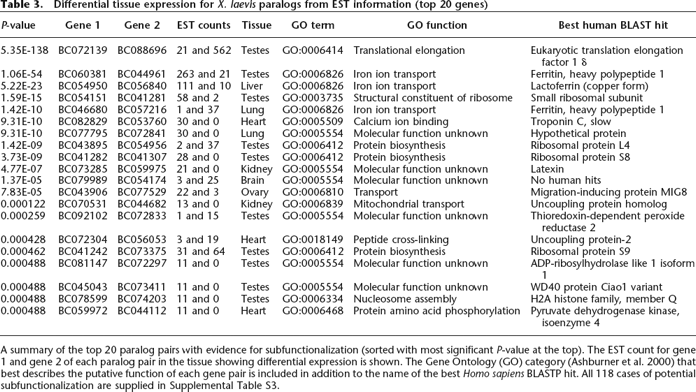
A summary of the top 20 paralog pairs with evidence for subfunctionalization (sorted with most significant P-value at the top). The EST count for gene 1 and gene 2 of each paralog pair in the tissue showing differential expression is shown. The Gene Ontology (GO) category (Ashburner et al. 2000) that best describes the putative function of each gene pair is included in addition to the name of the best Homo sapiens BLASTP hit. All 118 cases of potential subfunctionalization are supplied in Supplemental Table S3.











