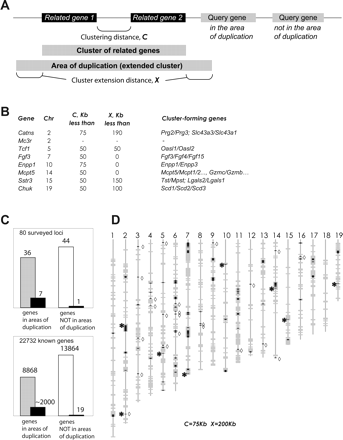
Asynchronously replicated genes are associated with regions of tandem gene duplication. (A) Defining the areas of gene duplication. Two or more genes are considered clustered if they encode related proteins (relatedness of proteins determined by BLASTP analysis of the complete nonredundant set of mouse protein-coding genes; see details in the text) and are no more than C Kb apart in the genome. To define an area of gene duplication, such a cluster is extended by X Kb in either direction. A gene is counted as belonging to an area of duplication if it overlaps in whole or partially with such an area; in the hypothetical depicted example, three genes fall within an area of gene duplication, and one does not. (B) Determining the values of parameters C and X for the whole-genome analysis. Minimal values of these parameters for each of the novel asynchronously replicated genes are shown. When the clustering parameters are set at C = 75 Kb, X = 200 Kb, then all these loci, with the exception of Mc3r, fall within the gene duplication areas. (C) Summary of the whole-genome clustering analysis. (Top) Of the 80 loci assessed in this study, 36 were in the areas of gene duplication (gray bar), and the rest were outside of such areas (white bar). Of the genes we identified as asynchronously replicated (black bars), seven were in the areas of duplication. (Bottom) 39% of mouse genes (8868 of 22,732 sequences analyzed) are in the areas of gene duplication (gray bar), with clustering parameters set at C = 75 Kb and X = 200 Kb uniformly applied to the locations of genes on the mouse genome assembly (NCBI build 33). The rest of the genes were not in such areas (white bar). Note that of known and presumed genes with asynchronous replication or random monoallelic expression (black bars) a vast majority are in the areas of gene duplication. (D) Distribution of the areas of gene duplication on the mouse genome. Locations of these areas were plotted on the mouse genome assembly (gray bars; acrosomes at top). A complete set of coordinates is in Supplemental Table S2. The duplicated areas largely overlap with the known or presumed loci with asynchronous replication (black tick marks): chemosensory receptor genes and pseudogenes, and known asynchronously replicated, monoallelically expressed genes from the immune system, as well as loci identified in this study as asynchronously replicated (asterisk to the left of the locus). Of ∼2000 known or presumed asynchronously replicated loci in the genome, we found only 19 that are apparent exceptions to this rule (diamond marks to the right of the locus).











