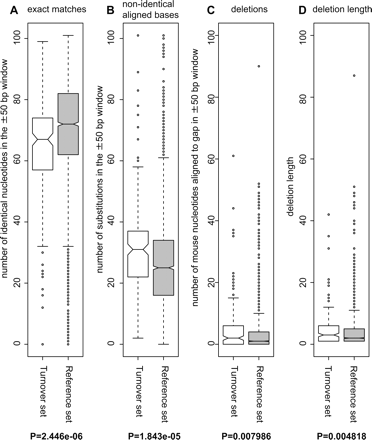
Sequence conservation is significantly lower in TSS regions with high levels of turnover. The degree of conservation in the genomic regions surrounding TSS (±50 bp) with high levels of turnover was investigated by using mouse/human BLASTZ NET alignments. Boxplots representing the (A) number of identical aligned nucleotides, (B) number of aligned nonidentical bases, and (C) number of deletions in the human genome using the mouse sequence as reference are shown for TSS regions exhibiting turnover and TSS regions from the reference set. (D) Distribution of lengths of deletions in the alignments. P-values resulting from a Wilcoxon two-tailed rank test between the sample and reference vectors are shown for each case.











