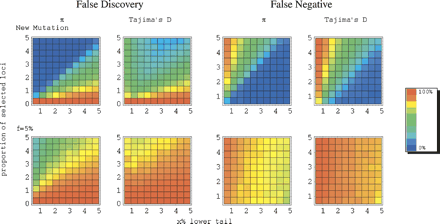
An estimate of error rates using π and Tajima’s D under the model for maize. Our aim was to mimic the approach of considering the lower tail of the empirical distribution of a summary statistic as significant. To do so, we generated 100 simulated data sets of 1000 loci, some fraction of which were linked to a selected site (see Methods for details). Shown are estimates of the proportion of loci in the tail that are neutrally evolving (i.e., the false-discovery rate) and the proportion of targets of selection not in the tail (i.e., the false-negative rate). Two selection models are considered (from top to bottom): (1) selection acted on a new mutation; (2) selection acted on a previously neutral allele and f = 5%. On the x-axis is the cutoff for significance, that is, the percentile of the distribution considered. On the y-axis is the proportion of loci linked to a target of selection in our simulated data set of 1000 loci.











