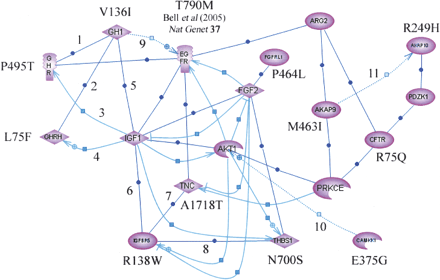
Inter-relationship between genes involved in the GH-IGF pathway containing SNPs associated with risk of lung cancer. Interactions were established using Pathway Assist software and are color-coded as follows: blue (expression), gray (regulation), and red (protein binding). Supporting publications are indicated with the corresponding NCBI Entrez PubMed ID in square brackets. (1) Binding [12,888,636]; (2) Binding [11,832,396]; (3) Expression [11,849,991]; (4) Expression [11,606,442]; (5) Binding [11,126,270]; (6) Binding [15,140,223]; (7) Binding [10,982,804]; (8) Binding [11,751,588]; (9) Regulation [14,517,795]; (10) Regulation [11,395,482]; (11) Regulation [15,047,863]. Validated nsSNPs with frequency data from Caucasian populations were not available in dbSNP Build 123 for genes AKT1, ARG2, FGF2, IGF1, PZDK1, and PRKCE. Bell et al. (2005) found association between lung cancer and SNP T790M.











