Effects of masking out pseudogenes
Click on table to view larger version.
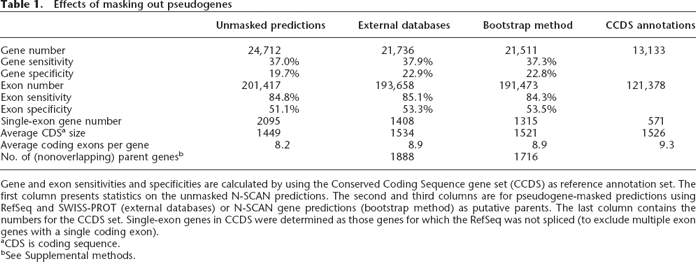
Gene and exon sensitivities and specificities are calculated by using the Conserved Coding Sequence gene set (CCDS) as reference annotation set. The first column presents statistics on the unmasked N-SCAN predictions. The second and third columns are for pseudogene-masked predictions using RefSeq and SWISS-PROT (external databases) or N-SCAN gene predictions (bootstrap method) as putative parents. The last column contains the numbers for the CCDS set. Single-exon genes in CCDS were determined as those genes for which the RefSeq was not spliced (to exclude multiple exon genes with a single coding exon).
aCDS is coding sequence.
bSee Supplemental methods.











