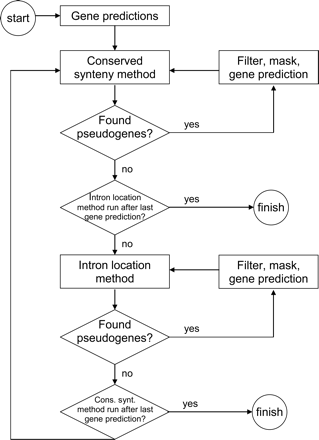
Flow diagram for the bootstrap method that combines pseudogene finding with gene prediction. To iteratively mask pseudogenes and rerun gene prediction, PPFINDER is run with a masking step after each of the methods (conserved synteny and intron alignment). This nested looping is done to remove redundancy, because many pseudogenes will be found by both methods. First, the cycle of pseudogene finding and masking is run using the conserved synteny method, until no more pseudogenes are found. Then the same is done using the intron alignment method. PPFINDER will keep looping through both methods until neither finds any more pseudogenes. One masking/gene prediction loop is called one round.











