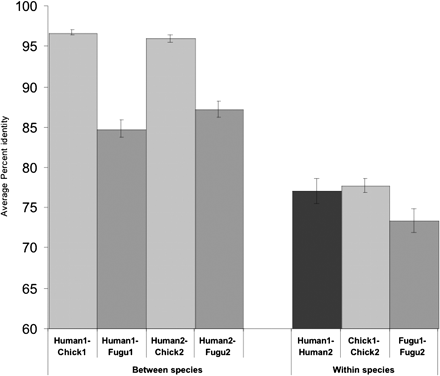
Mean percent sequence identities of related dCNEs within and between species. “Between species” represents orthologous dCNEs; dCNEs from two-member families are extremely well conserved between human and chicken copies (Human1–Chick1, Human2–Chick2) with a lower level of conservation between human and Fugu copies (Human1–Fugu1, Human2–Fugu2), reflecting the longer phylogenetic branch length and higher rate of evolution in fish genomes (Jaillon et al. 2004). Error bars represent the standard error of the mean. “Within species” represents dCNEs within the same genome; mean conservation is much lower between dCNEs within the same species than between orthologs, indicating an increased rate of evolution following duplication followed by extreme evolutionary constraint sometime prior to the fish–tetrapod divergence. For >80% of families that contained at least two members in Fugu, phylogenetic trees constructed using maximum parsimony (with 1000 bootstrap replicates) fitted the expected topology, i.e., dCNE family members were more similar between genomes than within genomes.











