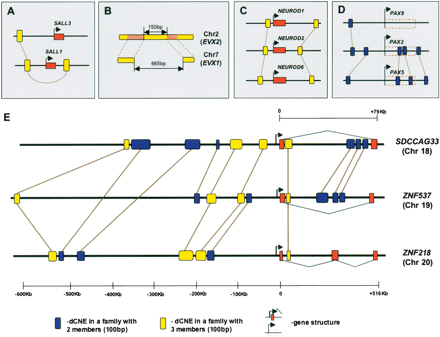
dCNE families with more than two members. Brown lines connect dCNEs within the same family. (A) An unusual three-member family is found around SALL1 and SALL3. Here, two of the members are found both 5′ and 3′ of SALL1, a feature not seen in any of the other families. (B) A three-member family of interest is located around EVX1 and EVX2. Here, the two members on Chr7 show significant similarity to different parts of the single element on Chr2 and are separated by a gap of 665 bp, little of which is conserved across orthologous regions in other vertebrates. The same region is only 150 bp on Chr2 and is conserved across vertebrates, indicating that this is likely to be the ancestral element. (C) dCNEs around NEUROD 1, 2, and 6 are retained in a similar manner to those in E although this set of paralogs contains no two-member families. (D) In contrast to dCNEs retained across three-member paralogous gene families as in C and E, PAX2, PAX5, and PAX8 retain only two-member dCNE families, connected by a central gene (PAX2). Blue boxes within the red dashed box represent dCNE located within the introns of these genes. (E) Four three-member families (yellow boxes) are located around three teashirt orthologs on human chromosomes 18, 19, and 20 that possess overlapping expression domains (Caubit et al. 2005). Additionally, seven two-member families (blue boxes) are retained between different pairs of these paralogs. Element lengths are represented relative to a 100-bp element shown in the key. Gene annotation was taken from Ensembl v27.35.1 for SDCCAG33 and ZNF537 and the Vertebrate Genome Annotation Database (http://vega.sanger.ac.uk/Homo_sapiens) for ZNF218. Distance of dCNEs from the presumed translation start site (TSS) in all three genes is fixed according to the lower scale. Different scales are used for the distance downstream of the TSS for ZNF218 (lower scale) and SDCCAG33 and ZNF537 (upper scale).











