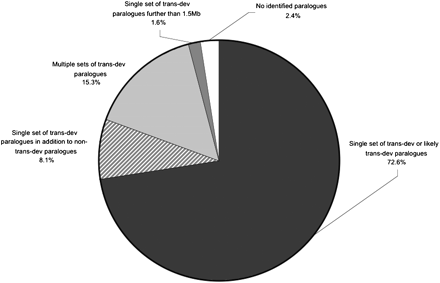
Presence of trans-dev paralogs in the vicinity of the 124 dCNE families. For the majority of families, trans-dev paralogs were detected within 1.5 Mb, either upstream or downstream of dCNEs. In most cases, just a single set of paralogs was detected with annotation relating to trans-dev (black), with some regions containing additional non trans-dev paralogs (striped). Some regions contained multiple sets of trans-dev paralogs (light gray). For dCNEs located in gene deserts, a search region up to the next known gene was used (dark gray). A small proportion of dCNEs were located in regions with no functionally annotated paralogs (white).











