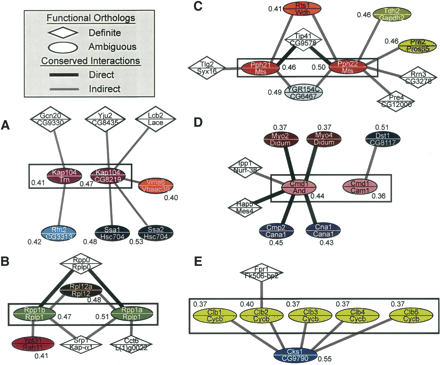
Example orthologs resolved by network conservation. Each node represents a putative functional match between a yeast/fly protein pair (with names shown above/below the line, respectively). Links between nodes denote conserved interactions (thick black, direct interactions in both species; thin gray, indirect interaction in one of the species; see Methods). Diamond- vs. oval-shaped nodes represent definite vs. ambiguous functional orthologs. Oval nodes of the same color represent ambiguous protein pairs belonging to the same Inparanoid cluster. The mean probability of functional orthology is given next to each ambiguous pair. Cluster 246 (A), 1439 (B), 211 (C), 917 (D), and 1104 (E) show examples of clusters that were disambiguated by conserved network information; the cluster resolved in each panel is outlined by a black rectangle.











