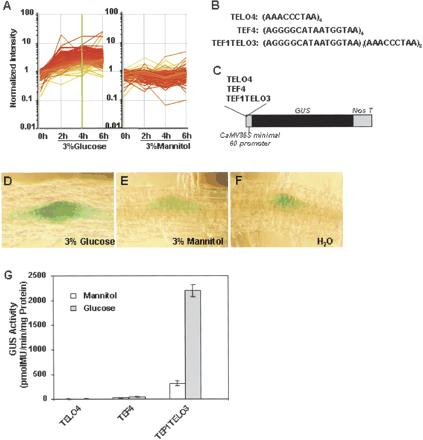
The TELO motif confers glucose-mediated transcriptional regulation. (A) Expression patterns of the glucose-up-regulated genes with promoters containing the TELO motif. (B) Sequences of the TELO4, TEF4, and TEF1TELO3 motifs. (C) Constructs containing the TELO4, TEF4, and TEF1TELO3 motifs in a –60 CaMV::GUS reporter vector are shown. An oligonucleotide tetramer of TELO (TELO4) and TEF (TEF4) motifs and a combined motif (TEF1TELO3) containing one TEF sequence and three TELO sequences were inserted upstream of the –60 CaMV::GUS reporter construct. (D,E,F) Histochemical analysis of GUS activity of TEF1TELO3::GUS transgenic plants in response to 3% glucose (D), 3% mannitol (E), and water (F) for 12 h. GUS activities in lateral root primordia are shown. (G) GUS activity of TEF4::GUS, TELO4::GUS, and TEF1TELO3::GUS transgenic plants. Protoplasts made from 7-d-old TEF4::GUS, TELO4::GUS, and TEF1TELO3::GUS transgenic plants were cultured in 400 mM glucose or 400 mM mannitol for 48 h before GUS activity was measured. Error bars represent the standard error of the mean from five samples. These transgenic lines were assayed at least three times.











